| Title: | Applies Display Metadata to Analysis Results Datasets |
|---|---|
| Description: | Creates a framework to store and apply display metadata to Analysis Results Datasets (ARDs). The use of 'tfrmt' allows users to define table format and styling without the data, and later apply the format to the data. |
| Authors: | Becca Krouse [aut, cre],
Christina Fillmore [aut] |
| Maintainer: | Becca Krouse <[email protected]> |
| License: | Apache License (>= 2) |
| Version: | 0.1.3.9000 |
| Built: | 2025-03-09 05:27:56 UTC |
| Source: | https://github.com/GSK-Biostatistics/tfrmt |
Apply formatting
apply_frmt(frmt_def, .data, value, mock = FALSE, ...) ## S3 method for class 'frmt' apply_frmt(frmt_def, .data, value, mock = FALSE, ...) ## S3 method for class 'frmt_combine' apply_frmt( frmt_def, .data, value, mock = FALSE, param, column, label, group, ... ) ## S3 method for class 'frmt_when' apply_frmt(frmt_def, .data, value, mock = FALSE, ...)apply_frmt(frmt_def, .data, value, mock = FALSE, ...) ## S3 method for class 'frmt' apply_frmt(frmt_def, .data, value, mock = FALSE, ...) ## S3 method for class 'frmt_combine' apply_frmt( frmt_def, .data, value, mock = FALSE, param, column, label, group, ... ) ## S3 method for class 'frmt_when' apply_frmt(frmt_def, .data, value, mock = FALSE, ...)
frmt_def |
formatting to be applied |
.data |
data, but only what is getting changed |
value |
value symbol should only be one |
mock |
Logical value is this is for a mock or not. By default |
... |
additional arguments for methods |
param |
param column as a quosure |
column |
column columns as a list of quosures |
label |
label column as a quosure |
group |
group column as a list of quosures |
formatted dataset
library(tibble) library(dplyr) # Set up data df <- tibble(x = c(20.12,34.54,12.34)) apply_frmt( frmt_def = frmt("XX.X"), .data=df, value=quo(x))library(tibble) library(dplyr) # Set up data df <- tibble(x = c(20.12,34.54,12.34)) apply_frmt( frmt_def = frmt("XX.X"), .data=df, value=quo(x))
Big N structure allows you to specify which values should become the subject totals ("big N" values)
and how they should be formatted in the table's column labels. Values are specified by providing
the value(s) of the param column for which the values are big N's.
This will remove these from the body of the table and place them into columns
matching the values in the column column(s). The default formatting is N = xx,
on its own line, but that can be changed by providing a different frmt() to
n_frmt
big_n_structure(param_val, n_frmt = frmt("\nN = xx"), by_page = FALSE)big_n_structure(param_val, n_frmt = frmt("\nN = xx"), by_page = FALSE)
param_val |
row value(s) of the parameter column for which the values are big n's |
n_frmt |
|
by_page |
Option to include different big Ns for each group-defined set of pages (defined by any variables set to ".default" in the |
big_n_structure object
Define the formatting of the body contents of the table through a series of frmt_structures. Structures get applied in order from bottom up, so the last added structure is the first applied.
body_plan(...)body_plan(...)
... |
list of frmt_structures defining the body formatting |
body_plan object
frmt_structure() defines which rows the formats will be applied
to, and frmt(), frmt_combine(), and frmt_when() define the format
semantics.
tfrmt_spec<- tfrmt( title = "Table Title", body_plan = body_plan( frmt_structure( group_val = c("group1"), label_val = ".default", frmt("XXX") ) ) )tfrmt_spec<- tfrmt( title = "Table Title", body_plan = body_plan( frmt_structure( group_val = c("group1"), label_val = ".default", frmt("XXX") ) ) )
Using <tidy-select> expressions and a series
span_structures, define the order of the columns. The selection follows "last
selected" principals, meaning columns are moved to the last selection as
opposed to preserving the first location.
col_plan(..., .drop = FALSE) span_structure(...)col_plan(..., .drop = FALSE) span_structure(...)
... |
For a col_plan and span_structure,
< |
.drop |
Boolean. Should un-listed columns be dropped from the data. Defaults to FALSE. |
When col_plan gets applied and is used to create the output table, the underlying logic sorts out which column specifically is being selected. If a column is selected twice, the last instance in which the column is selected will be the location it gets rendered.
Avoid beginning the col_plan() column selection with a deselection (i.e.
col_plan(-col1), col_plan(-starts_with("value"))). This will
result in the table preserving all columns not "de-selected" in the
statement, and the order of the columns not changed. It is preferred when
creating the col_plan() to identify all the columns planned on
preserving in the order they are wished to appear, or if
<tidy-select> arguments - such as
everything- are used, identify the de-selection after
the positive-selection.
Alternatively, once the gt table is produced, use the
cols_hide function to remove un-wanted columns.
col_plan object
span_structure object
Here are some example outputs:

library(dplyr) ## select col_1 as the first column, remove col_last, then create spanning ## structures that have multiple levels ## ## examples also assume the tfrmt has the column argument set to c(c1, c2, c3) ## spanning_col_plan_ex <- col_plan( col_1, -col_last, span_structure( c1 = "Top Label Level 1", c2 = "Second Label Level 1.1", c3 = c(col_3, col_4) ), span_structure( c1 = "Top Label Level 1", c2 = "Second Label Level 1.2", c3 = starts_with("B") ), span_structure( c1 = "Top Label Level 1", c3 = col_5 ), span_structure( c2 = "Top Label Level 2", c3 = c(col_6, col_7) ) ) ## select my_col_1 as the first column, then ## rename col_2 to new_col_1 and put as the ## second column, then select the rest of the columns renaming_col_plan_ex <- col_plan( my_col_1, new_col_1 = col_2, everything() ) renaming_col_plan_ex2 <- col_plan( my_col_1, new_col_1 = col_2, span_structure( c1 = c(`My Favorite span name` = "Top Label Level 1"), c3 = c(`the results column` = col_5) ) )library(dplyr) ## select col_1 as the first column, remove col_last, then create spanning ## structures that have multiple levels ## ## examples also assume the tfrmt has the column argument set to c(c1, c2, c3) ## spanning_col_plan_ex <- col_plan( col_1, -col_last, span_structure( c1 = "Top Label Level 1", c2 = "Second Label Level 1.1", c3 = c(col_3, col_4) ), span_structure( c1 = "Top Label Level 1", c2 = "Second Label Level 1.2", c3 = starts_with("B") ), span_structure( c1 = "Top Label Level 1", c3 = col_5 ), span_structure( c2 = "Top Label Level 2", c3 = c(col_6, col_7) ) ) ## select my_col_1 as the first column, then ## rename col_2 to new_col_1 and put as the ## second column, then select the rest of the columns renaming_col_plan_ex <- col_plan( my_col_1, new_col_1 = col_2, everything() ) renaming_col_plan_ex2 <- col_plan( my_col_1, new_col_1 = col_2, span_structure( c1 = c(`My Favorite span name` = "Top Label Level 1"), c3 = c(`the results column` = col_5) ) )
Define how the columns of the table body should be aligned, whether left, right or on a specific character(s).
col_style_plan(...)col_style_plan(...)
... |
series of col_style_structure objects |
col_style_plan object
col_style_structure() for more information on how to specify how to and which columns to align.
plan <- col_style_plan( col_style_structure(col = "my_var", align = "left", width = 100), col_style_structure(col = vars(four), align = "right"), col_style_structure(col = vars(two, three), align = c(".", ",", " ")) )plan <- col_style_plan( col_style_structure(col = "my_var", align = "left", width = 100), col_style_structure(col = vars(four), align = "right"), col_style_structure(col = vars(two, three), align = c(".", ",", " ")) )
Column Style Structure
col_style_structure( col, align = NULL, type = c("char", "pos"), width = NULL, ... )col_style_structure( col, align = NULL, type = c("char", "pos"), width = NULL, ... )
col |
Column value to align on from |
align |
Alignment to be applied to column. Defaults to |
type |
Type of alignment: "char" or "pos", for character alignment (default), and positional alignment, respectively. Positional alignment allows for aligning over multiple positions in the column. |
width |
Width to apply to the column in number of characters. Acceptable values include a numeric value, or a character string of a number. |
... |
These dots are for future extensions and must be empty |
Supports alignment and width setting of data value columns (values found in the column column). Row group and label columns are left-aligned by default. Acceptable input values for align differ by type = "char" or "pos":
"left" for left alignment
"right" for right alignment"
supply a vector of character(s) to align on. If more than one character is provided, alignment will be based on the first occurrence of any of the characters. For alignment based on white space, leading white spaces will be ignored.
supply a vector of strings covering all formatted cell values, with numeric values represented as x's. These values can be created manually or obtained by utilizing the helper display_val_frmts(). Alignment positions will be represented by vertical bars. For example, with starting values: c("12.3", "(5%)", "2.35 (10.23)") we can align all of the first sets of decimals and parentheses by providing align = c("xx|.x", "||(x%)", "x|.xx |")
col_style_structure object
col_style_plan() for more information on how to combine
col_style_structure()'s together to form a plan.
plan <- col_style_plan( col_style_structure(col = "my_var", align = c("xx| |(xx%)", "xx|.x |(xx.x - xx.x)"), type = "pos", width = 100), col_style_structure(col = vars(four), align = "right", width = 200), col_style_structure(col = vars(two, three), align = c(".", ",", " ")), col_style_structure(col = c(two, three), width = 25), col_style_structure(col = two, width = 25), col_style_structure(col = span_structure(span = value, col = val2), width = 25) )plan <- col_style_plan( col_style_structure(col = "my_var", align = c("xx| |(xx%)", "xx|.x |(xx.x - xx.x)"), type = "pos", width = 100), col_style_structure(col = vars(four), align = "right", width = 200), col_style_structure(col = vars(two, three), align = c(".", ",", " ")), col_style_structure(col = c(two, three), width = 25), col_style_structure(col = two, width = 25), col_style_structure(col = span_structure(span = value, col = val2), width = 25) )
A dataset containing the results needed for an AE table. Using the CDISC pilot data.
data_aedata_ae
A data frame with 2,794 rows and 8 variables:
highest level row labels: System Organ Class
more specific row labels: Preferred Term
higher level column names (spanners)
lower level column names
parameter to explain each value
values to put in a table
controls ordering
more ordering controls
A dataset containing the results needed for a demography table. Using the CDISC pilot data.
data_demogdata_demog
A data frame with 386 rows and 7 variables:
highest level row labels
more specific row labels
parameter to explain each value
grouping column used to distinguish continuous and categorical
controls ordering
more ordering controls
column names
values to put in a table
A dataset containing the results needed for an Efficacy table. Using the CDISC pilot data for ADAS-Cog(11).
data_efficacydata_efficacy
A data frame with 70 rows and 7 variables:
highest level row labels
more specific row labels
column names
parameter to explain each value
values to put in a table
controls ordering
more ordering controls
A dataset containing the results needed for an labs results table. Using the CDISC pilot data.
data_labsdata_labs
A data frame with 4,950 rows and 7 variables:
highest level row labels: Lab value class
more specific row labels: Lab parameter
most specific row labels: Study visit
higher level column names (spanners)
lower level column names
parameter to explain each value
values to put in a table
controls ordering
more ordering controls
more ordering controls
Used when debugging formatting, it is an easy way to allow you to see which formats are applied to each row in your dataset.
Used when debugging formatting, it is an easy way to allow you to see which formats are applied to each row in your dataset.
display_row_frmts(tfrmt, .data, convert_to_txt = TRUE) display_row_frmts(tfrmt, .data, convert_to_txt = TRUE)display_row_frmts(tfrmt, .data, convert_to_txt = TRUE) display_row_frmts(tfrmt, .data, convert_to_txt = TRUE)
tfrmt |
tfrmt object to apply to the data |
.data |
Data to apply the tfrmt to |
convert_to_txt |
Logical value converting formatting to text, by default
|
formatted tibble
formatted tibble
library(dplyr) library(tidyr) tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value, body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt_combine( "{count} {percent}", count = frmt("xxx"), percent = frmt_when("==100"~ frmt(""), "==0"~ "", "TRUE" ~ frmt("(xx.x%)")))) )) # Create data df <- crossing(label = c("label 1", "label 2"), column = c("placebo", "trt1"), param = c("count", "percent")) %>% mutate(value=c(24,19,2400/48,1900/38,5,1,500/48,100/38)) display_row_frmts(tfrmt_spec,df) library(dplyr) library(tidyr) tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value, body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt_combine( "{count} {percent}", count = frmt("xxx"), percent = frmt_when("==100"~ frmt(""), "==0"~ "", "TRUE" ~ frmt("(xx.x%)")))) )) # Create data df <- crossing(label = c("label 1", "label 2"), column = c("placebo", "trt1"), param = c("count", "percent")) %>% mutate(value=c(24,19,2400/48,1900/38,5,1,500/48,100/38)) display_row_frmts(tfrmt_spec,df)library(dplyr) library(tidyr) tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value, body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt_combine( "{count} {percent}", count = frmt("xxx"), percent = frmt_when("==100"~ frmt(""), "==0"~ "", "TRUE" ~ frmt("(xx.x%)")))) )) # Create data df <- crossing(label = c("label 1", "label 2"), column = c("placebo", "trt1"), param = c("count", "percent")) %>% mutate(value=c(24,19,2400/48,1900/38,5,1,500/48,100/38)) display_row_frmts(tfrmt_spec,df) library(dplyr) library(tidyr) tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value, body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt_combine( "{count} {percent}", count = frmt("xxx"), percent = frmt_when("==100"~ frmt(""), "==0"~ "", "TRUE" ~ frmt("(xx.x%)")))) )) # Create data df <- crossing(label = c("label 1", "label 2"), column = c("placebo", "trt1"), param = c("count", "percent")) %>% mutate(value=c(24,19,2400/48,1900/38,5,1,500/48,100/38)) display_row_frmts(tfrmt_spec,df)
A helper for creating positional-alignment specifications for the col_style_plan. Returns all unique formatted values to appear in the column(s) specified. Numeric values are represented by x's.
display_val_frmts(tfrmt, .data, mock = FALSE, col = NULL)display_val_frmts(tfrmt, .data, mock = FALSE, col = NULL)
tfrmt |
tfrmt object to apply to the data |
.data |
Data to apply the tfrmt to |
mock |
Mock table? TRUE or FALSE (default) |
col |
Column value to align on from |
text representing character vector of formatted values to be copied and modified in the col_style_plan
tf_spec <- tfrmt( group = c(rowlbl1,grp), label = rowlbl2, column = column, param = param, value = value, sorting_cols = c(ord1, ord2), body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt_combine("{n} ({pct} %)", n = frmt("xxx"), pct = frmt("xx.x"))), frmt_structure(group_val = ".default", label_val = "n", frmt("xxx")), frmt_structure(group_val = ".default", label_val = c("Mean", "Median", "Min","Max"), frmt("xxx.x")), frmt_structure(group_val = ".default", label_val = "SD", frmt("xxx.xx")), frmt_structure(group_val = ".default", label_val = ".default", p = frmt_when(">0.99" ~ ">0.99", "<0.15" ~ "<0.15", TRUE ~ frmt("x.xxx", missing = ""))) )) display_val_frmts(tf_spec, data_demog, col = vars(everything())) display_val_frmts(tf_spec, data_demog, col = "p-value")tf_spec <- tfrmt( group = c(rowlbl1,grp), label = rowlbl2, column = column, param = param, value = value, sorting_cols = c(ord1, ord2), body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt_combine("{n} ({pct} %)", n = frmt("xxx"), pct = frmt("xx.x"))), frmt_structure(group_val = ".default", label_val = "n", frmt("xxx")), frmt_structure(group_val = ".default", label_val = c("Mean", "Median", "Min","Max"), frmt("xxx.x")), frmt_structure(group_val = ".default", label_val = "SD", frmt("xxx.xx")), frmt_structure(group_val = ".default", label_val = ".default", p = frmt_when(">0.99" ~ ">0.99", "<0.15" ~ "<0.15", TRUE ~ frmt("x.xxx", missing = ""))) )) display_val_frmts(tf_spec, data_demog, col = vars(everything())) display_val_frmts(tf_spec, data_demog, col = "p-value")
Element block
element_block(post_space = c(NULL, " ", "-"), fill = TRUE)element_block(post_space = c(NULL, " ", "-"), fill = TRUE)
post_space |
Values to show in a new line created after the group block |
fill |
Whether to recycle the value of |
element block object
row_grp_plan() for more details on how to group row group
structures, row_grp_structure() for more details on how to specify row group
structures, element_row_grp_loc() for more details on how to
specify whether row group titles span the entire table or collapse.
tfrmt_spec <- tfrmt( group = grp1, label = label, param = param, value = value, column = column, row_grp_plan = row_grp_plan( row_grp_structure(group_val = ".default", element_block(post_space = " ")) ), body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt("xx")) ) )tfrmt_spec <- tfrmt( group = grp1, label = label, param = param, value = value, column = column, row_grp_plan = row_grp_plan( row_grp_structure(group_val = ".default", element_block(post_space = " ")) ), body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt("xx")) ) )
Element Row Group Location
element_row_grp_loc( location = c("indented", "spanning", "column", "noprint", "gtdefault"), indent = " " )element_row_grp_loc( location = c("indented", "spanning", "column", "noprint", "gtdefault"), indent = " " )
location |
Location of the row group labels. Specifying 'indented' combines all group and label variables into a single column with each sub-group indented under its parent. 'spanning' and 'column' retain the highest level group variable in its own column and combine all remaining group and label variables into a single column with sub-groups indented. The highest level group column will either be printed as a spanning header or in its own column in the gt. The 'noprint' option allows the user to suppress group values from being printed. Finally, the 'gtdefault' option allows users to use the 'gt' defaults for styling multiple group columns. |
indent |
A string of the number of spaces you want to indent |
element_row_grp_loc object
Here are some example outputs:

row_grp_plan() for more details on how to group row group
structures, row_grp_structure() for more details on how to specify row
group structures, element_block() for more details on how to specify
spacing between each group.
tfrmt_spec <- tfrmt( group = c(grp1, grp2), label = label, param = param, value = value, column = column, row_grp_plan = row_grp_plan(label_loc = element_row_grp_loc(location = "noprint")), body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt("xx")) ) )tfrmt_spec <- tfrmt( group = c(grp1, grp2), label = label, param = param, value = value, column = column, row_grp_plan = row_grp_plan(label_loc = element_row_grp_loc(location = "noprint")), body_plan = body_plan( frmt_structure(group_val = ".default", label_val = ".default", frmt("xx")) ) )
Defining the location and content of footnotes with a series of footnote structures. Each structure is a footnote and can be applied in multiple locations.
footnote_plan(..., marks = c("numbers", "letters", "standard", "extended"))footnote_plan(..., marks = c("numbers", "letters", "standard", "extended"))
... |
a series of |
marks |
type of marks required for footnotes, properties inherited from tab_footnote in 'gt'. Available options are "numbers", "letters", "standard" and "extended" (standard for a traditional set of 4 symbols, extended for 6 symbols). The default option is set to "numbers". |
footnote plan object
# Adds a footnote indicated by letters rather than numbers to Group 1 footnote_plan <- footnote_plan( footnote_structure(footnote_text = "Source Note", group_val = "Group 1"), marks="letters") # Adds a footnote to the 'Placebo' column footnote_plan <- footnote_plan( footnote_structure(footnote_text = "footnote", column_val = "Placebo"), marks="numbers")# Adds a footnote indicated by letters rather than numbers to Group 1 footnote_plan <- footnote_plan( footnote_structure(footnote_text = "Source Note", group_val = "Group 1"), marks="letters") # Adds a footnote to the 'Placebo' column footnote_plan <- footnote_plan( footnote_structure(footnote_text = "footnote", column_val = "Placebo"), marks="numbers")
Footnote Structure
footnote_structure( footnote_text, column_val = NULL, group_val = NULL, label_val = NULL )footnote_structure( footnote_text, column_val = NULL, group_val = NULL, label_val = NULL )
footnote_text |
string with text for footnote |
column_val |
string or a named list of strings which represent the column to apply the footnote to |
group_val |
string or a named list of strings which represent the value of group to apply the footnote to |
label_val |
string which represents the value of label to apply the footnote to |
footnote structure object
# Adds a source note aka a footnote without a symbol in the table footnote_structure <- footnote_structure(footnote_text = "Source Note") # Adds a footnote to the 'Placebo' column footnote_structure <- footnote_structure(footnote_text = "Text", column_val = "Placebo") # Adds a footnote to either 'Placebo' or 'Treatment groups' depending on which # which is last to appear in the column vector footnote_structure <- footnote_structure(footnote_text = "Text", column_val = list(col1 = "Placebo", col2= "Treatment groups")) # Adds a footnote to the 'Adverse Event' label footnote_structure <- footnote_structure("Text", label_val = "Adverse Event")# Adds a source note aka a footnote without a symbol in the table footnote_structure <- footnote_structure(footnote_text = "Source Note") # Adds a footnote to the 'Placebo' column footnote_structure <- footnote_structure(footnote_text = "Text", column_val = "Placebo") # Adds a footnote to either 'Placebo' or 'Treatment groups' depending on which # which is last to appear in the column vector footnote_structure <- footnote_structure(footnote_text = "Text", column_val = list(col1 = "Placebo", col2= "Treatment groups")) # Adds a footnote to the 'Adverse Event' label footnote_structure <- footnote_structure("Text", label_val = "Adverse Event")
These functions provide an abstracted way to approach to define formatting of table contents. By defining in this way, the formats can be layered to be more specific and general cell styling can be done first.
frmt() is the base definition of a format. This defines spacing, rounding,
and missing behaviour.
frmt_combine() is used when two or more rows need to be combined into a
single cell in the table. Each of the rows needs to have a defined frmt()
and need to share a label.
frmt_when() is used when a rows format behaviour is dependent on the value itself and is written similarly to dplyr::case_when().
The left hand side of the equation is a "TRUE"for the default case or the right hand side of a boolean expression ">50".
frmt(expression, missing = NULL, scientific = NULL, transform = NULL, ...) frmt_combine(expression, ..., missing = NULL) frmt_when(..., missing = NULL)frmt(expression, missing = NULL, scientific = NULL, transform = NULL, ...) frmt_combine(expression, ..., missing = NULL) frmt_when(..., missing = NULL)
expression |
this is the string representing the intended format. See details: expression for more a detailed description. |
missing |
when a value is missing that is intended to be formatted, what value to place. See details: missing for more a detailed description. |
scientific |
a string representing the intended scientific notation to be appended to the expression. Ex. "e^XX" or " x10^XX". |
transform |
this is what should happen to the value prior to formatting,
It should be a formula or function. Ex. |
... |
See details: |
frmt() All numbers are represented by "x". Any additional character are
printed as-is. If additional X's present to the left of the decimal point
than the value, they will be represented as spaces.
frmt_combine() defines how the parameters will be combined as a
glue::glue() statement. Parameters need to be equal to the values in the
param column and defined in the expression as "{param1} {param2}".
frmt() Value to enter when the value is missing. When NULL, the value
is "".
frmt_combine() defines how when all values to be combined are missing.
When NULL the value is "".
frmt() These dots are for future extensions and must be
empty.
frmt_combine() accepts named arguments defining the frmt() to
be applied to which parameters before being combined.
frmt_when()accepts a series of equations separated by commas, similar
to dplyr::case_when(). The left hand side of the equation is a "TRUE"for the
default case or the right hand side of a boolean expression ">50". The
right hand side of the equation is the frmt() to apply when the left
side evaluates to TRUE.
frmt object
body_plan() combines the frmt_structures to be applied to the
table body, and frmt_structure() defines which rows the formats will be applied
to.
frmt("XXX %") frmt("XX.XXX") frmt("xx.xx", scientific = "x10^xx") frmt_combine( "{param1} {param2}", param1 = frmt("XXX %"), param2 = frmt("XX.XXX") ) frmt_when( ">3" ~ frmt("(X.X%)"), "<=3" ~ frmt("Undetectable") ) frmt_when( "==100"~ frmt(""), "==0"~ "", "TRUE" ~ frmt("(XXX.X%)") )frmt("XXX %") frmt("XX.XXX") frmt("xx.xx", scientific = "x10^xx") frmt_combine( "{param1} {param2}", param1 = frmt("XXX %"), param2 = frmt("XX.XXX") ) frmt_when( ">3" ~ frmt("(X.X%)"), "<=3" ~ frmt("Undetectable") ) frmt_when( "==100"~ frmt(""), "==0"~ "", "TRUE" ~ frmt("(XXX.X%)") )
Function needed to create a frmt_structure object, which is a building block
of body_plan(). This specifies the rows the format will be applied to.
frmt_structure(group_val = ".default", label_val = ".default", ...)frmt_structure(group_val = ".default", label_val = ".default", ...)
group_val |
A string or a named list of strings which represent the value of group should be when the given frmt is implemented |
label_val |
A string which represent the value of label should be when the given frmt is implemented |
... |
either a |
frmt_structure object
Here are some example outputs:

body_plan() combines the frmt_structures to be applied to the
table body, and frmt(), frmt_combine(), and frmt_when() define the
format semantics.
sample_structure <- frmt_structure( group_val = c("group1"), label_val = ".default", frmt("XXX") ) ## multiple group columns sample_structure <- frmt_structure( group_val = list(grp_col1 = "group1", grp_col2 = "subgroup3"), label_val = ".default", frmt("XXX") )sample_structure <- frmt_structure( group_val = c("group1"), label_val = ".default", frmt("XXX") ) ## multiple group columns sample_structure <- frmt_structure( group_val = list(grp_col1 = "group1", grp_col2 = "subgroup3"), label_val = ".default", frmt("XXX") )
Check if input is a frmt
Check if input is a frmt_combine
Check if input is a frmt_when
Check if input is a frmt_structure
Check if input is a row_grp_structure
is_frmt(x) is_frmt_combine(x) is_frmt_when(x) is_frmt_structure(x) is_row_grp_structure(x)is_frmt(x) is_frmt_combine(x) is_frmt_when(x) is_frmt_structure(x) is_row_grp_structure(x)
x |
Object to check |
'TRUE' if yes, 'FALSE' if no
x1 <- frmt("XXX.XX") is_frmt(x1) x2 <- frmt_combine("XXX %","XX,XXX") is_frmt_combine(x2) x2 <- frmt_when( ">3" ~ frmt("(X.X%)"), "<=3" ~ frmt("Undetectable") ) is_frmt_when(x2) x3 <- frmt_structure( group_val = c("group1"), label_val = ".default", frmt("XXX") ) is_frmt_structure(x3) x4 <- row_grp_structure(group_val = c("A","C"), element_block(post_space = "---")) is_row_grp_structure(x4)x1 <- frmt("XXX.XX") is_frmt(x1) x2 <- frmt_combine("XXX %","XX,XXX") is_frmt_combine(x2) x2 <- frmt_when( ">3" ~ frmt("(X.X%)"), "<=3" ~ frmt("Undetectable") ) is_frmt_when(x2) x3 <- frmt_structure( group_val = c("group1"), label_val = ".default", frmt("XXX") ) is_frmt_structure(x3) x4 <- row_grp_structure(group_val = c("A","C"), element_block(post_space = "---")) is_row_grp_structure(x4)
Reader to read JSON files/objects into tfrmt objects
json_to_tfrmt(path = NULL, json = NULL)json_to_tfrmt(path = NULL, json = NULL)
path |
location of the json file to read in |
json |
json object to read in. By default this is null. This function will read in json object preferentially. So if both a path and a json object are supplied the json object will be read in. |
Provide utility for layering tfrmt objects together. If both tfrmt's have values, it will preferentially choose the second tfrmt by default. This is an alternative to piping together tfrmt's
layer_tfrmt(x, y, ..., join_body_plans = TRUE)layer_tfrmt(x, y, ..., join_body_plans = TRUE)
x, y
|
tfrmt objects that need to be combined |
... |
arguments passed to layer_tfrmt_arg functions for combining different tfrmt elements |
join_body_plans |
should the |
When combining two body_plans, the body plans will stack together, first the body plan from x tfrmt then y tfrmt. This means that frmt_structures in y will take priority over those in x.
Combining two tfrmt with large body_plans can lead to slow table evaluation.
Consider setting join_body_plan to FALSE. Only the y body_plan will be
preserved.
tfrmt object
tfrmt_1 <- tfrmt(title = "title1") tfrmt_2 <- tfrmt(title = "title2",subtitle = "subtitle2") layered_table_format <- layer_tfrmt(tfrmt_1, tfrmt_2)tfrmt_1 <- tfrmt(title = "title1") tfrmt_2 <- tfrmt(title = "title2",subtitle = "subtitle2") layered_table_format <- layer_tfrmt(tfrmt_1, tfrmt_2)
Make mock data for display shells
make_mock_data(tfrmt, .default = 1:3, n_cols = NULL)make_mock_data(tfrmt, .default = 1:3, n_cols = NULL)
tfrmt |
tfrmt object |
.default |
Number of unique levels to create for group/label values set to ".default" |
n_cols |
Number of columns in the output table (not including
group/label variables). If not supplied it will default to using the
|
tibble containing mock data
tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value, body_plan = body_plan( frmt_structure(group_val=".default", label_val=".default", frmt("xx.x")) ) ) make_mock_data(tfrmt_spec)tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value, body_plan = body_plan( frmt_structure(group_val=".default", label_val=".default", frmt("xx.x")) ) ) make_mock_data(tfrmt_spec)
Defining the location and/or frequency of page splits with a series of page_structure's and the row_every_n argument, respectively.
page_plan( ..., note_loc = c("noprint", "preheader", "subtitle", "source_note"), max_rows = NULL )page_plan( ..., note_loc = c("noprint", "preheader", "subtitle", "source_note"), max_rows = NULL )
... |
a series of |
note_loc |
Location of the note describing each table's subset value(s).
Useful if the |
max_rows |
Option to set a maximum number of rows per page. Takes a numeric value. |
page_plan object
# use of page_struct page_plan( page_structure(group_val = "grp1", label_val = "lbl1") ) # use of # rows page_plan( max_rows = 5 )# use of page_struct page_plan( page_structure(group_val = "grp1", label_val = "lbl1") ) # use of # rows page_plan( max_rows = 5 )
Page structure
page_structure(group_val = NULL, label_val = NULL)page_structure(group_val = NULL, label_val = NULL)
group_val |
string or a named list of strings which represent the value of group to split after. Set to ".default" if the split should occur after every unique value of the variable. |
label_val |
string which represents the value of label to split after. Set to ".default" if the split should occur after every unique value of the variable. |
page structure object
# split page after every unique level of the grouping variable page_structure(group_val = ".default", label_val = NULL) # split page after specific levels page_structure(group_val = "grp1", label_val = "lbl3")# split page after every unique level of the grouping variable page_structure(group_val = ".default", label_val = NULL) # split page after specific levels page_structure(group_val = "grp1", label_val = "lbl3")
Set custom parameter-level significant digits rounding
param_set(...)param_set(...)
... |
Series of name-value pairs, optionally formatted using
|
Type param_set() in console to view package defaults. Use of the
function will add to the defaults and/or override included defaults of the
same name. For values that are integers, use NA so no decimal places will
be added.
list of default parameter-level significant digits rounding
# View included defaults param_set() # Update the defaults param_set("{mean} ({sd})" = c(2,3), "pct" = 1) # Separate mean and SD to different lines param_set("mean" = 2, "sd" = 3) # Add formatting using the glue syntax param_set("{pct} %" = 1)# View included defaults param_set() # Update the defaults param_set("{mean} ({sd})" = c(2,3), "pct" = 1) # Separate mean and SD to different lines param_set("mean" = 2, "sd" = 3) # Add formatting using the glue syntax param_set("{pct} %" = 1)
Print mock table to GT
print_mock_gt( tfrmt, .data = NULL, .default = 1:3, n_cols = NULL, .unicode_ws = TRUE )print_mock_gt( tfrmt, .data = NULL, .default = 1:3, n_cols = NULL, .unicode_ws = TRUE )
tfrmt |
tfrmt the mock table will be based off of |
.data |
Optional data. If this is missing, group values, labels values and parameter values will be estimated based on the tfrmt |
.default |
sequence to replace the default values if a dataset isn't provided |
n_cols |
the number of columns. This will only be used if mock data isn't
provided. If not supplied, it will default to using the |
.unicode_ws |
Whether to convert white space to unicode in preparation for output |
a stylized gt object
# Create tfrmt specification
tfrmt_spec <- tfrmt( label = label, column =
column, param = param, body_plan = body_plan( frmt_structure(group_val =
".default", label_val = ".default", frmt_combine( "{count} {percent}",
count = frmt("xxx"), percent = frmt_when("==100"~ frmt(""), "==0"~ "",
"TRUE" ~ frmt("(xx.x%)")))) ))
# Print mock table using default
print_mock_gt(tfrmt = tfrmt_spec)

# Create mock data
df <- crossing(label = c("label 1", "label 2",
"label 3"), column = c("placebo", "trt1", "trt2"), param = c("count",
"percent"))
# Print mock table using mock data
print_mock_gt(tfrmt_spec, df)

Print to ggplot
print_to_ggplot(tfrmt, .data, ...)print_to_ggplot(tfrmt, .data, ...)
tfrmt |
tfrmt object that will dictate the structure of the ggplot object |
.data |
Data to style in order to make the ggplot object |
... |
Inputs to geom_text to modify the style of the table body |
a stylized ggplot object
# Create data
risk<-tibble(time=c(rep(c(0,1000,2000,3000),3)),
label=c(rep("Obs",4),rep("Lev",4),rep("Lev+5FU",4)),
value=c(630,372,256,11,620,360,266,8,608,425,328,14),
param=rep("n",12))
table<-tfrmt(
label = label ,
column = time,
param = param,
value = value) %>%
print_to_ggplot(risk)
table

Print to gt
print_to_gt(tfrmt, .data, .unicode_ws = TRUE)print_to_gt(tfrmt, .data, .unicode_ws = TRUE)
tfrmt |
tfrmt object that will dictate the structure of the table |
.data |
Data to style in order to make the table |
.unicode_ws |
Whether to convert white space to unicode in preparation for output |
a stylized gt object
library(dplyr)
# Create tfrmt specification
tfrmt_spec <- tfrmt(
label = label,
column = column,
param = param,
value=value,
body_plan = body_plan(
frmt_structure(group_val = ".default", label_val = ".default",
frmt_combine(
"{count} {percent}",
count = frmt("xxx"),
percent = frmt_when("==100"~ frmt(""),
"==0"~ "",
"TRUE" ~ frmt("(xx.x%)"))))
))
# Create data
df <- crossing(label = c("label 1", "label 2"),
column = c("placebo", "trt1"),
param = c("count", "percent")) %>%
mutate(value=c(24,19,2400/48,1900/38,5,1,500/48,100/38))
print_to_gt(tfrmt_spec,df)

Define the look of the table groups on the output. This function allows you to add spaces after blocks and allows you to control how the groups are viewed whether they span the entire table or are nested as a column.
row_grp_plan(..., label_loc = element_row_grp_loc(location = "indented"))row_grp_plan(..., label_loc = element_row_grp_loc(location = "indented"))
... |
Row group structure objects separated by commas |
label_loc |
|
row_grp_plan object
row_grp_structure() for more details on how to specify row group
structures, element_block() for more details on how to specify spacing
between each group, element_row_grp_loc() for more details on how to
specify whether row group titles span the entire table or collapse.
## single grouping variable example sample_grp_plan <- row_grp_plan( row_grp_structure(group_val = c("A","C"), element_block(post_space = "---")), row_grp_structure(group_val = c("B"), element_block(post_space = " ")), label_loc = element_row_grp_loc(location = "column") ) ## example with multiple grouping variables sample_grp_plan <- row_grp_plan( row_grp_structure(group_val = list(grp1 = "A", grp2 = "b"), element_block(post_space = " ")), label_loc = element_row_grp_loc(location = "spanning") )## single grouping variable example sample_grp_plan <- row_grp_plan( row_grp_structure(group_val = c("A","C"), element_block(post_space = "---")), row_grp_structure(group_val = c("B"), element_block(post_space = " ")), label_loc = element_row_grp_loc(location = "column") ) ## example with multiple grouping variables sample_grp_plan <- row_grp_plan( row_grp_structure(group_val = list(grp1 = "A", grp2 = "b"), element_block(post_space = " ")), label_loc = element_row_grp_loc(location = "spanning") )
Function needed to create a row_grp_structure object, which is a building block
of row_grp_plan()
row_grp_structure(group_val = ".default", element_block)row_grp_structure(group_val = ".default", element_block)
group_val |
A string or a named list of strings which represent the value of group should be when the given frmt is implemented |
element_block |
element_block() object to define the block styling |
row_grp_structure object
row_grp_plan() for more details on how to group row group
structures, element_block() for more details on how to specify spacing
between each group.
## single grouping variable example row_grp_structure(group_val = c("A","C"), element_block(post_space = "---")) ## example with multiple grouping variables row_grp_structure(group_val = list(grp1 = "A", grp2 = "b"), element_block(post_space = " "))## single grouping variable example row_grp_structure(group_val = c("A","C"), element_block(post_space = "---")) ## example with multiple grouping variables row_grp_structure(group_val = list(grp1 = "A", grp2 = "b"), element_block(post_space = " "))
tfrmt, or "table format" is a way to pre-define the non-data components of your tables, and how the data will be handled once added: i.e. title, footers, headers, span headers, and cell formats. In addition, tfrmt's can be layered, building from one table format to the next. For cases where only one value can be used, the newly defined tfrmt accepts the latest tfrmt
tfrmt( tfrmt_obj, group = vars(), label = quo(), param = quo(), value = quo(), column = vars(), title, subtitle, row_grp_plan, body_plan, col_style_plan, col_plan, sorting_cols, big_n, footnote_plan, page_plan, ... )tfrmt( tfrmt_obj, group = vars(), label = quo(), param = quo(), value = quo(), column = vars(), title, subtitle, row_grp_plan, body_plan, col_style_plan, col_plan, sorting_cols, big_n, footnote_plan, page_plan, ... )
tfrmt_obj |
a tfrmt object to base this new format off of |
group |
what are the grouping vars of the input dataset |
label |
what is the label column of the input dataset |
param |
what is the param column of the input dataset |
value |
what is the value column of the input dataset |
column |
what is the column names column in the input dataset |
title |
title of the table |
subtitle |
subtitle of the table |
row_grp_plan |
plan of the row groups blocking. Takes a |
body_plan |
combination and formatting of the input data. Takes a |
col_style_plan |
how to style columns including alignment (left, right, character) and width. Takes a |
col_plan |
a col_plan object which is used to select, rename, and nest
columns. Takes a |
sorting_cols |
which columns determine sorting of output |
big_n |
how to format subject totals ("big Ns") for inclusion in the column labels. Takes a |
footnote_plan |
footnotes to be added to the table. Takes a |
page_plan |
pagination splits to be applied to the table. Takes a |
... |
These dots are for future extensions and must be empty. |
tfrmt allows users to pass vars, quo, and unquoted expressions to a
variety of arguments, such as group, label, param, value,
column, and sorting_cols. Users accustomed to tidyverse semantics
should be familiar with this behaviour. However, there is an important
behaviour difference between tfrmt and normal tidyverse functions. Because
the data are not a part of tfrmt, it does not know when a value being
passed to it is intended to be an unquoted expression representing a column
name or an object from the environment. As such, it preferentially uses the
value from the environment over preserving the entry as an expression. For
example, if you have an object "my_object" in your
environment with the value "Hello world", and try to create a tfrmt as
tfrmt(column = my_object), it will take the value of "my_object" over
assuming the column argument is an unquoted expression and view the entry
to column as "Hello World". To pass "my_object" to tfrmt as a column name, use
quotes around the value: tfrmt(columnn = "my_object").
Additionally, unquoted expressions that match tfrmt's other
argument names can cause unexpected results. It is recommended
to put quotes around the value as such:
tfrmt(label = "group"). In this case, the quoting will prevent tfrmt
from assigning its group input value to the label value.
tfrmt object
Here are some example outputs:

tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value) tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value, # Set the formatting for values body_plan = body_plan( frmt_structure( group_val = ".default", label_val = ".default", frmt_combine("{n} {pct}", n = frmt("xxx"), pct = frmt_when( "==100" ~ "(100%)", "==0" ~ "", TRUE ~ frmt("(xx.x %)") ) ) ) ), # Specify column styling plan col_style_plan = col_style_plan( col_style_structure(col = vars(everything()), align = c(".",","," ")) )) tfrmt_spec <- tfrmt( group = group, label = label, column = column, param = param, value=value, sorting_cols = c(ord1, ord2), # specify value formatting body_plan = body_plan( frmt_structure( group_val = ".default", label_val = ".default", frmt_combine("{n} {pct}", n = frmt("xxx"), pct = frmt_when( "==100" ~ "(100%)", "==0" ~ "", TRUE ~ frmt("(xx.x %)") ) ) ), frmt_structure( group_val = ".default", label_val = "n", frmt("xxx") ), frmt_structure( group_val = ".default", label_val = c("Mean", "Median", "Min","Max"), frmt("xxx.x") ), frmt_structure( group_val = ".default", label_val = "SD", frmt("xxx.xx") ), frmt_structure( group_val = ".default", label_val = ".default", p = frmt("") ), frmt_structure( group_val = ".default", label_val = c("n","<65 yrs","<12 months","<25"), p = frmt_when( ">0.99" ~ ">0.99", "<0.001" ~ "<0.001", TRUE ~ frmt("x.xxx", missing = "") ) ) ), # remove extra cols col_plan = col_plan(-grp, -starts_with("ord") ), # Specify column styling plan col_style_plan = col_style_plan( col_style_structure(col = vars(everything()), align = c(".",","," ")) ), # Specify row group plan row_grp_plan = row_grp_plan( row_grp_structure( group_val = ".default", element_block(post_space = " ") ), label_loc = element_row_grp_loc(location = "column") ) )tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value) tfrmt_spec <- tfrmt( label = label, column = column, param = param, value=value, # Set the formatting for values body_plan = body_plan( frmt_structure( group_val = ".default", label_val = ".default", frmt_combine("{n} {pct}", n = frmt("xxx"), pct = frmt_when( "==100" ~ "(100%)", "==0" ~ "", TRUE ~ frmt("(xx.x %)") ) ) ) ), # Specify column styling plan col_style_plan = col_style_plan( col_style_structure(col = vars(everything()), align = c(".",","," ")) )) tfrmt_spec <- tfrmt( group = group, label = label, column = column, param = param, value=value, sorting_cols = c(ord1, ord2), # specify value formatting body_plan = body_plan( frmt_structure( group_val = ".default", label_val = ".default", frmt_combine("{n} {pct}", n = frmt("xxx"), pct = frmt_when( "==100" ~ "(100%)", "==0" ~ "", TRUE ~ frmt("(xx.x %)") ) ) ), frmt_structure( group_val = ".default", label_val = "n", frmt("xxx") ), frmt_structure( group_val = ".default", label_val = c("Mean", "Median", "Min","Max"), frmt("xxx.x") ), frmt_structure( group_val = ".default", label_val = "SD", frmt("xxx.xx") ), frmt_structure( group_val = ".default", label_val = ".default", p = frmt("") ), frmt_structure( group_val = ".default", label_val = c("n","<65 yrs","<12 months","<25"), p = frmt_when( ">0.99" ~ ">0.99", "<0.001" ~ "<0.001", TRUE ~ frmt("x.xxx", missing = "") ) ) ), # remove extra cols col_plan = col_plan(-grp, -starts_with("ord") ), # Specify column styling plan col_style_plan = col_style_plan( col_style_structure(col = vars(everything()), align = c(".",","," ")) ), # Specify row group plan row_grp_plan = row_grp_plan( row_grp_structure( group_val = ".default", element_block(post_space = " ") ), label_loc = element_row_grp_loc(location = "column") ) )
This function creates an tfrmt for an n % table, so count based table. The
parameter values for n and percent can be provided (by default it will assume
n and pct). Additionally the frmt_when for formatting the percent can
be specified. By default 100% and 0% will not appear and everything between
99% and 100% and 0% and 1% will be rounded using greater than (>) and less
than (<) signs respectively.
tfrmt_n_pct( n = "n", pct = "pct", pct_frmt_when = frmt_when("==100" ~ frmt(""), ">99" ~ frmt("(>99%)"), "==0" ~ "", "<1" ~ frmt("(<1%)"), "TRUE" ~ frmt("(xx.x%)")), tfrmt_obj = NULL )tfrmt_n_pct( n = "n", pct = "pct", pct_frmt_when = frmt_when("==100" ~ frmt(""), ">99" ~ frmt("(>99%)"), "==0" ~ "", "<1" ~ frmt("(<1%)"), "TRUE" ~ frmt("(xx.x%)")), tfrmt_obj = NULL )
n |
name of count (n) value in the parameter column |
pct |
name of percent (pct) value in the parameter column |
pct_frmt_when |
formatting to be used on the the percent values |
tfrmt_obj |
an optional tfrmt object to layer |
tfrmt object
print_mock_gt(tfrmt_n_pct())

This function creates a tfrmt based on significant digits specifications for
group/label values. The input data spec provided to sigdig_df will contain
group/label value specifications. tfrmt_sigdig assumes that these columns
are group columns unless otherwise specified. The user may optionally choose
to pass the names of the group and/or label columns as arguments to the
function.
tfrmt_sigdig( sigdig_df, group = vars(), label = quo(), param_defaults = param_set(), missing = NULL, tfrmt_obj = NULL, ... )tfrmt_sigdig( sigdig_df, group = vars(), label = quo(), param_defaults = param_set(), missing = NULL, tfrmt_obj = NULL, ... )
sigdig_df |
data frame containing significant digits formatting spec.
Has 1 record per group/label value, and columns for relevant group and/or
label variables, as well as a numeric column |
group |
what are the grouping vars of the input dataset |
label |
what is the label column of the input dataset |
param_defaults |
Option to override or add to default parameters. |
missing |
missing option to be included in all |
tfrmt_obj |
an optional tfrmt object to layer |
... |
These dots are for future extensions and must be empty. |
Currently covers specifications for frmt and
frmt_combine. frmt_when not supported and must be supplied in additional
tfrmt that is layered on.
If the group/label variables are not provided to the arguments, the body_plan will be constructed from the input data with the following behaviour:
If no group or label are supplied, it will be assumed that all columns in the input data are group columns.
If a label variable is provided, but nothing is
specified for group, any leftover columns (i.e. not matching sigdig or the
supplied label variable name) in the input data will be assumed to be group
columns.
If any group variable is provided, any leftover columns (i.e. not
matching sigdig or the supplied group/label variable) will be disregarded.
tfrmt object with a body_plan constructed based on the
significant digits data spec and param-level significant digits defaults.
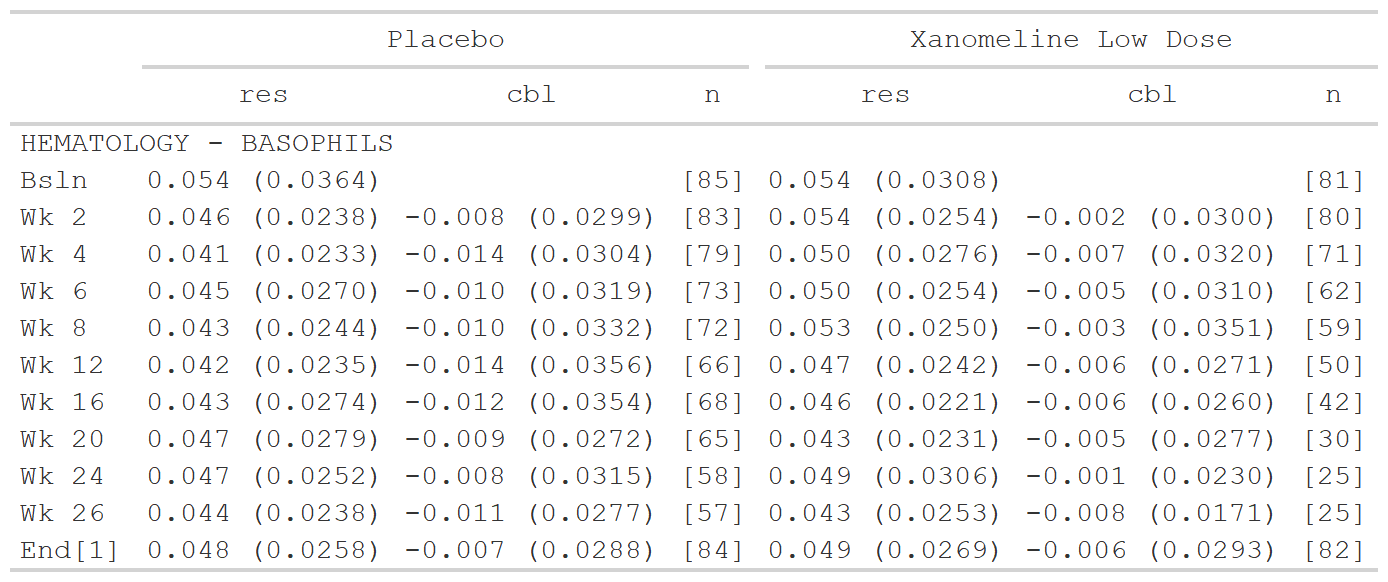
sig_input <- tibble::tribble(
~group1, ~group2, ~sigdig,
"CHEMISTRY", ".default", 3,
"CHEMISTRY", "ALBUMIN", 1,
"CHEMISTRY", "CALCIUM", 1,
".default", ".default", 2
)
# Subset data for the example
data <- dplyr::filter(data_labs, group2 == "BASOPHILS", col1 %in% c("Placebo", "Xanomeline Low Dose"))
tfrmt_sigdig(sigdig_df = sig_input,
group = vars(group1, group2),
label = rowlbl,
param_defaults = param_set("[{n}]" = NA)) %>%
tfrmt(column = vars(col1, col2),
param = param,
value = value,
sorting_cols = vars(ord1, ord2, ord3),
col_plan = col_plan(-starts_with("ord"))) %>%
print_to_gt(.data = data)
Print to JSON
tfrmt_to_json(tfrmt, path = NULL)tfrmt_to_json(tfrmt, path = NULL)
tfrmt |
tfrmt to print |
path |
file path to save JSON to. If not provided the JSON will just print to the console |
JSON
tfrmt( label = label, column = column, param = param, value=value) %>% tfrmt_to_json()tfrmt( label = label, column = column, param = param, value=value) %>% tfrmt_to_json()
Remap group values in a tfrmt
update_group(tfrmt, ...)update_group(tfrmt, ...)
tfrmt |
a |
... |
Use new_name = old_name to rename selected variables |
A tfrmt with the group variables updated in all places
tfrmt object with updated groups#'
tfrmt_spec <- tfrmt( group = c(group1, group2), body_plan = body_plan( frmt_structure( group_val = list(group2 = "value"), label_val = ".default", frmt("XXX") ), frmt_structure( group_val = list(group1 = "value", group2 = "value"), label_val = ".default", frmt("XXX") ) )) tfrmt_spec %>% update_group(New_Group = group1)tfrmt_spec <- tfrmt( group = c(group1, group2), body_plan = body_plan( frmt_structure( group_val = list(group2 = "value"), label_val = ".default", frmt("XXX") ), frmt_structure( group_val = list(group1 = "value", group2 = "value"), label_val = ".default", frmt("XXX") ) )) tfrmt_spec %>% update_group(New_Group = group1)